Classification: inference
In this notebook we demonstrate how to use the script scripts/classification/main_classification_inference.py to identify organisms in clips to taxonomic groups.
We first import the necessary packages, and specify the path where our main scripts are found:
[6]:
import os, sys
from pathlib import Path
import yaml
from mzbsuite.utils import cfg_to_arguments
# !pip install -e ../
sys.path.append("../scripts/")
The most important function in this task is inference_classifier, and we specifically import it here for ease:
[7]:
# from classification.main_classification_finetune import main as finetune_classifier
from classification.main_classification_inference import main as inference_classifier
?inference_classifier
Signature: inference_classifier(args, cfg)
Docstring:
Function to run inference on macrozoobenthos images clips, using a trained model.
Parameters
----------
args : argparse.Namespace
Namespace containing the arguments passed to the script. Notably:
- input_dir: path to the directory containing the images to be classified
- input_model: path to the directory containing the model to be used for inference
- output_dir: path to the directory where the results will be saved
- config_file: path to the config file with train / inference parameters
cfg : dict
Dictionary containing the configuration parameters.
Returns
-------
None. Saves the results in the specified folder.
File: /data/users/luca/mzb-workflow/scripts/classification/main_classification_inference.py
Type: function
Next we declare the running parameters, telling the script where to find the input data, the model and where to put the outputs. In this notebook, we pass these as a Python dictionary, but a shell (.sh) achieving the same result can be found in workflows/run_pipeline.sh.
[8]:
ROOT_DIR = Path.cwd()
print(ROOT_DIR)
MODEL = "efficientnet-b2-v0"
arguments = {
"config_file": ROOT_DIR.parent.absolute() / "configs/configuration_flume_datasets.yaml",
"input_dir": ROOT_DIR.parent.absolute() / "data/mzb_example_data/training_dataset/val_set/",
"input_model": ROOT_DIR.parent.absolute() / f"models/mzb-classification-models/{MODEL}",
"output_dir": ROOT_DIR.parent.absolute() / "results/mzb_example_data/classification",
}
with open(str(arguments["config_file"]), "r") as f:
cfg = yaml.load(f, Loader=yaml.FullLoader)
cfg["trcl_gpu_ids"] = None
cfg
/data/users/luca/mzb-workflow/notebooks
[8]:
{'glob_random_seed': 222,
'glob_root_folder': '/data/users/luca/mzb-workflow/mzb-workflow/',
'glob_blobs_folder': '/data/users/luca/mzb-workflow/mzb-workflow/data/derived/blobs/',
'glob_local_format': 'pdf',
'model_logger': 'wandb',
'impa_image_format': 'jpg',
'impa_clip_areas': [2750, 4900],
'impa_area_threshold': 5000,
'impa_gaussian_blur': [21, 21],
'impa_gaussian_blur_passes': 3,
'impa_adaptive_threshold_block_size': 351,
'impa_mask_postprocess_kernel': [11, 11],
'impa_mask_postprocess_passes': 5,
'impa_bounding_box_buffer': 200,
'impa_save_clips_plus_features': True,
'lset_class_cut': 'order',
'lset_val_size': 0.1,
'lset_taxonomy': '/data/users/luca/mzb-workflow/data/MZB_taxonomy.csv',
'trcl_learning_rate': 0.001,
'trcl_batch_size': 16,
'trcl_weight_decay': 0,
'trcl_step_size_decay': 5,
'trcl_number_epochs': 10,
'trcl_save_topk': 1,
'trcl_num_classes': 8,
'trcl_model_pretrarch': 'efficientnet-b2',
'trcl_num_workers': 16,
'trcl_wandb_project_name': 'mzb-classifiers',
'trcl_logger': 'wandb',
'trsk_learning_rate': 0.001,
'trsk_batch_size': 32,
'trsk_weight_decay': 0,
'trsk_step_size_decay': 25,
'trsk_number_epochs': 400,
'trsk_save_topk': 1,
'trsk_num_classes': 2,
'trsk_model_pretrarch': 'mit_b2',
'trsk_num_workers': 16,
'trsk_wandb_project_name': 'mzb-skeletons',
'trsk_logger': 'wandb',
'infe_model_ckpt': 'last',
'infe_num_classes': 8,
'infe_image_glob': '*_rgb.jpg',
'skel_class_exclude': 'errors',
'skel_conv_rate': 131.6625,
'skel_label_thickness': 3,
'skel_label_buffer_on_preds': 25,
'skel_label_clip_with_mask': False,
'trcl_gpu_ids': None}
We now need to transform parameters in the configuration file in a format that Python can understand (a dictionary in this case), we can use the provided helper function cfg_to_arguments:
[9]:
# Transforms configurations dicts to argparse arguments
args_p = cfg_to_arguments(arguments)
cfg_p = cfg_to_arguments(cfg)
print(str(cfg_p))
{'glob_random_seed': 222, 'glob_root_folder': '/data/users/luca/mzb-workflow/mzb-workflow/', 'glob_blobs_folder': '/data/users/luca/mzb-workflow/mzb-workflow/data/derived/blobs/', 'glob_local_format': 'pdf', 'model_logger': 'wandb', 'impa_image_format': 'jpg', 'impa_clip_areas': [2750, 4900], 'impa_area_threshold': 5000, 'impa_gaussian_blur': [21, 21], 'impa_gaussian_blur_passes': 3, 'impa_adaptive_threshold_block_size': 351, 'impa_mask_postprocess_kernel': [11, 11], 'impa_mask_postprocess_passes': 5, 'impa_bounding_box_buffer': 200, 'impa_save_clips_plus_features': True, 'lset_class_cut': 'order', 'lset_val_size': 0.1, 'lset_taxonomy': '/data/users/luca/mzb-workflow/data/MZB_taxonomy.csv', 'trcl_learning_rate': 0.001, 'trcl_batch_size': 16, 'trcl_weight_decay': 0, 'trcl_step_size_decay': 5, 'trcl_number_epochs': 10, 'trcl_save_topk': 1, 'trcl_num_classes': 8, 'trcl_model_pretrarch': 'efficientnet-b2', 'trcl_num_workers': 16, 'trcl_wandb_project_name': 'mzb-classifiers', 'trcl_logger': 'wandb', 'trsk_learning_rate': 0.001, 'trsk_batch_size': 32, 'trsk_weight_decay': 0, 'trsk_step_size_decay': 25, 'trsk_number_epochs': 400, 'trsk_save_topk': 1, 'trsk_num_classes': 2, 'trsk_model_pretrarch': 'mit_b2', 'trsk_num_workers': 16, 'trsk_wandb_project_name': 'mzb-skeletons', 'trsk_logger': 'wandb', 'infe_model_ckpt': 'last', 'infe_num_classes': 8, 'infe_image_glob': '*_rgb.jpg', 'skel_class_exclude': 'errors', 'skel_conv_rate': 131.6625, 'skel_label_thickness': 3, 'skel_label_buffer_on_preds': 25, 'skel_label_clip_with_mask': False, 'trcl_gpu_ids': None}
Once we have all set up, we can simply call the inference_classifier function, which will use the provided parameters to produce predictions and a summary of classification accuracy (against a val set if it is provided).
[10]:
inference_classifier(args_p, cfg_p)
GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
IPU available: False, using: 0 IPUs
HPU available: False, using: 0 HPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0,1]
Predicting DataLoader 0: 100%|██████████| 4/4 [00:00<00:00, 9.33it/s]
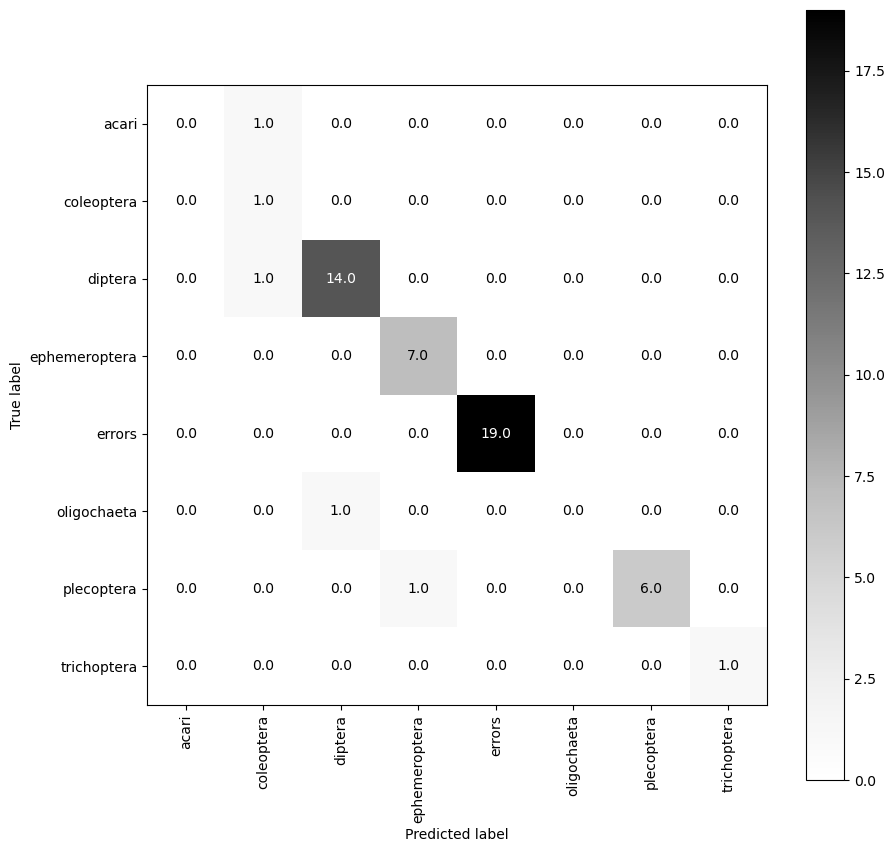
The produced confusion matrix show how many predictions the model made that corresponded to the real identity of the organism (on the diagonal), and also when the model was wrong what it guessed instead (off diagonal).